Difference between revisions of "Blast2go"
| Line 32: | Line 32: | ||
'''Steps to follow for a very large fasta file''' | '''Steps to follow for a very large fasta file''' | ||
| − | 1) Split into N number of files using python script | + | You will probably wish to do this in your scratch directory |
| + | |||
| + | 1) Split into N number of files using python script | ||
[mjv08@bert ~]$ python fastaSplitByNumSubsets.py | [mjv08@bert ~]$ python fastaSplitByNumSubsets.py | ||
Split fasta file into a number of subsets | Split fasta file into a number of subsets | ||
USAGE: $0 <fasta file> <num of subsets> <prefix for subsets> | USAGE: $0 <fasta file> <num of subsets> <prefix for subsets> | ||
| + | |||
| + | e.g. | ||
| + | |||
| + | [mjv08@bert ~]$ python fastaSplitByNumSubsets.py file.fasta 300 split | ||
| + | |||
| + | 2) Blast your split file. You will probably need to write a slightly more complicated SGE script to do this; | ||
| + | |||
| + | #$ -S /bin/sh | ||
| + | |||
| + | #$ -q amd.q | ||
| + | #$ -cwd | ||
| + | #$ -l h_vmem=20G,h_stack=2G | ||
| + | |||
| + | module load BLAST | ||
| + | |||
| + | echo blastx -db /ibers/ernie/scratch/databases/db/nr -query $FILENAME -out $FILENAME".xml" -evalue 1e-3 -outfmt 5 -show_gis | sh | ||
Revision as of 11:22, 16 December 2013
Running blast2go
We have a blast2go server accessible within the Aberystwyth University network. You need to run the blast2go java program located here;
You can select the amount of memory you wish to use on your own machine by selecting the value in the download box. Note, you will require java installed on your machine. This can be downloaded from [java.com].
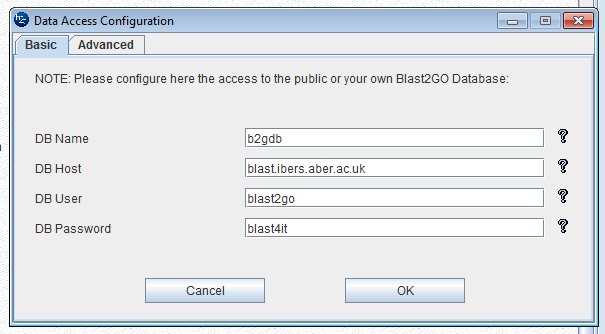
The, you will run the blast2go download. In Tools >> General Settings >> DataAccess Settings you can enter the details of the local server;
Running blast2go from the HPC command line
THIS IS WORK IN PROGRESS AND IS NOT TESTED. THIS IS JUST COMPILED NOTES FROM VARIOUS SOURCES IN ORDER TO TEST THE SEQUENCE. - mjv08 - 26-07-13
This assumes you have a fasta file and you wish to recreate the default setting of blast2go. This would include the blast, mapping and annotation steps with default blast2go settings.
Let's blast it;
blastall -p blastx -d nr -i myseqs.fasta -e 0.001 -m 7 -o blastResult.xml -I T -v 20 -b 20 -F L
where myseqs.fasta is the file you're blasting and blastResults.xml is your blasted results.
NOTE: According to the literature it says 'Note: Do not create xml-files with more than 100 xml-results. Pasrsing them will otherwise be difficult'
The next bit is to run the blast2go pipeline, which is the command line version;
java -Xms256m -Xmx512m -jar blast2go.jar -in blastResult.xml -v -a -out MyAnnot -d MyAnnot.dat -p b2gPipe.properties
Steps to follow for a very large fasta file
You will probably wish to do this in your scratch directory
1) Split into N number of files using python script
[mjv08@bert ~]$ python fastaSplitByNumSubsets.py Split fasta file into a number of subsets USAGE: $0 <fasta file> <num of subsets> <prefix for subsets>
e.g.
[mjv08@bert ~]$ python fastaSplitByNumSubsets.py file.fasta 300 split
2) Blast your split file. You will probably need to write a slightly more complicated SGE script to do this;
#$ -S /bin/sh
#$ -q amd.q #$ -cwd #$ -l h_vmem=20G,h_stack=2G
module load BLAST
echo blastx -db /ibers/ernie/scratch/databases/db/nr -query $FILENAME -out $FILENAME".xml" -evalue 1e-3 -outfmt 5 -show_gis | sh