Blast2go
Running blast2go
We have a blast2go server accessible within the Aberystwyth University network. You need to run the blast2go java program located here;
You can select the amount of memory you wish to use on your own machine by selecting the value in the download box. Note, you will require java installed on your machine. This can be downloaded from [java.com].
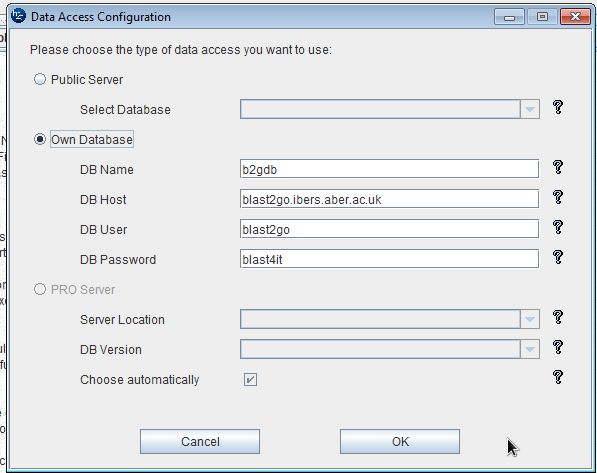
The, you will run the blast2go download. In Tools >> General Settings >> DataAccess Settings you can enter the details of the local server;
Running blast2go from the HPC command line
THIS IS WORK IN PROGRESS AND IS NOT TESTED. THIS IS JUST COMPILED NOTES FROM VARIOUS SOURCES IN ORDER TO TEST THE SEQUENCE. - mjv08 - 26-07-13
This assumes you have a fasta file and you wish to recreate the default setting of blast2go. This would include the blast, mapping and annotation steps with default blast2go settings.
Let's blast it;
blastall -p blastx -d nr -i myseqs.fasta -e 0.001 -m 7 -o blastResult.xml -I T -v 20 -b 20 -F L
where myseqs.fasta is the file you're blasting and blastResults.xml is your blasted results.
NOTE: According to the literature it says 'Note: Do not create xml-files with more than 100 xml-results. Pasrsing them will otherwise be difficult'
The next bit is to run the blast2go pipeline, which is the command line version;
java -Xms256m -Xmx512m -jar blast2go.jar -in blastResult.xml -v -a -out MyAnnot -d MyAnnot.dat -p b2gPipe.properties
Steps to follow for a very large fasta file
You will probably wish to do this in your scratch directory. REMEMBER! Scratch isn't backed up, so once you have something you wish to keep. Compress it and copy/move it to your home directory or an appropriate repository.
1) Split into N number of files using python script
[mjv08@bert ~]$ python fastaSplitByNumSubsets.py Split fasta file into a number of subsets USAGE: $0 <fasta file> <num of subsets> <prefix for subsets>
e.g.
[mjv08@bert ~]$ python fastaSplitByNumSubsets.py file.fasta 300 split
2) Blast your split file. You will probably need to write a slightly more complicated SGE script to do this;
#$ -S /bin/sh
#$ -q amd.q #$ -cwd #$ -l h_vmem=20G,h_stack=2G
module load BLAST
echo blastx -db /ibers/ernie/scratch/databases/db/nr -query $FILENAME -out $FILENAME".xml" -evalue 1e-3 -outfmt 5 -show_gis | sh
This is a bit more complicated that usual because this script takes in the file name as a parameter and then runs it. This is important because if you have 300 files you want to blast, you don't want to have to write 300 SGE scripts.
To run this script, you would type the following;
[mjv08@bert ~]$ qsub -N friendly_name -V filename_NUM.fasta mysgescript.sge
Obviously you would also have to run this for each of your split fasta files, replacing the filename_NUM.fasta with the name of your fasta file.
NOTE: To make things traceable, I would advise you to also set the friendly name to the same name as your fasta file. This will make it easier to trace errors.
Okay, so if you have lots of split fasta files and you don't want to type them all in to submit them. You can do the following;
[mjv08@bert ~]$ for i in `ls -1 split.*.fasta`; do qsub -N $i -V $i mysgescript.sge; done
HOWEVER!!!! Be very careful with this. It first assumes that your files are labeled;
split.1.fasta split.2.fasta
etc
It is also a good idea to add an echo before the qsub in order to see what is going to be submitted. e.g.
[mjv08@bert ~]$ for i in `ls -1 split.*.fasta`; do echo qsub -N $i -V $i mysgescript.sge; done
Also, you may wish to test it just on one or two first to see if you have set it up correctly. This can be achieved by piping it to head. e.g.
[mjv08@bert ~]$ for i in `ls -1 split.*.fasta`; do qsub -N $i -V $i mysgescript.sge; done | head
This will run only a few and you can see if it's working correctly.
3) Okay, blast2go step.
Download blast2go pipe (http://www.blast2go.com/b2glaunch/resources)
From the command line you can do the following;
[mjv08@bert ~] wget http://www.blast2go.com/data/blast2go/b2g4pipe_v2.5.zip .
and then;
[mjv08@bert ~] unzip b2g4pipe_v2.5.zip
for ease of use, it's best to work within the b2g4pipe directory. This is the line to run the blast2go pipe on a xml output from blast. You will probably need to load java as a module too.
[mjv08@bert ~] module load java [mjv08@bert ~] java -Xms3048m -Xmx3049m -cp *:ext/*: es.blast2go.prog.B2GAnnotPipe -in split_1_fasta.xml -out results/split_1_fasta.xml_out -prop b2gPipe.properties -v -annot -dat -img -annex -goslim -wiki html_template.html
but obviously this will run on bert, which is not ideal. We want it on the nodes.