Difference between revisions of "Running BLAST optimally"
| Line 8: | Line 8: | ||
[[File:Amd test.png]] | [[File:Amd test.png]] | ||
| + | |||
| + | This shows that increasing the number of threads adds around 2-3GB vmem per thread after the initial ~10GB vmem used for 1 thread. There is an oddity when using 2 and 3 threads where the vmem usage seems to decrease. | ||
Revision as of 14:23, 7 August 2013
This is a brief description of a couple of experiments that I (mjv08) have tried out using blast in order to determine the best way in which to run it.
Okay, so you have a fasta file you wish to blast. The file I had was supplied by Russ (rom) and contained 18788 sequences in a fasta file. He wanted to run the default settings for blast using blast2go. These settings using blast+ on the command line are;
blastx -db nr -query input.fasta -out output.xml -evalue 1e-3 -outfmt 5 -show_gis -num_threads 1
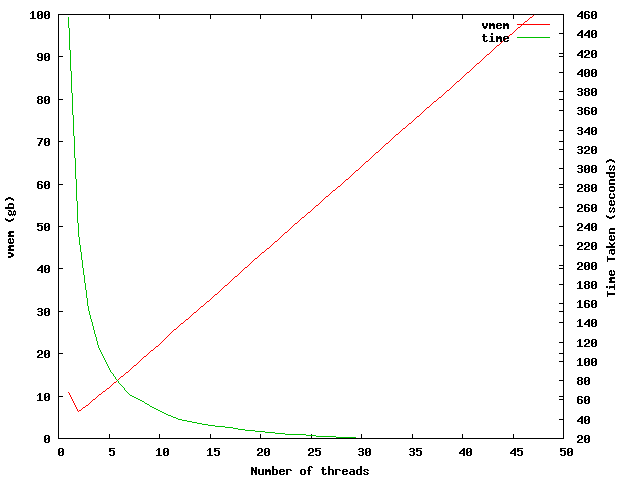
The first thing I have done is to take a single sequence (length=998) and used blastx against the ncbi nr database. This was run on a 32core AMD node and increased the number of threads that blast uses. The graph below shows the results of this, with the time taken and vmem used to complete the job.
This shows that increasing the number of threads adds around 2-3GB vmem per thread after the initial ~10GB vmem used for 1 thread. There is an oddity when using 2 and 3 threads where the vmem usage seems to decrease.